Movement through Bottlenecks#
This notebook can be directly downloaded here to run it locally.
In this following, we’ll investigate the movement of a crowd through two successive bottlenecks with a simulation. We expect that a jam occurs at the first bottleneck but not at the second one since the flow is considerably reduced by the first bottleneck.
For this purpose, we’ll setup a simulation scenario according to the RiMEA Test 12 [1] and analyse the results with pedpy to inspect the density and flow. After that we’ll vary the width of the bottlenecks and investigate the effects on the movement.
Let’s begin by importing the required packages for our simulation:
import pathlib
import jupedsim as jps
import matplotlib.pyplot as plt
import pandas as pd
import pedpy
from numpy.random import normal # normal distribution of free movement speed
from shapely import GeometryCollection, Polygon
Geometry Setup#
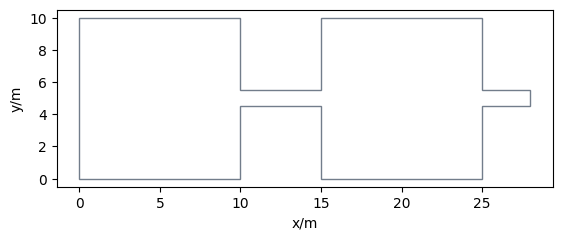
Let’s construct the geometry according to RiMEA by defining two rooms and a corridor with a width of 1 meter. To consider the interactions of agents in the second bottleneck (when leaving the second room) the corridor ends 3 meters behind the second room. By creating the union of all parts we end up with the whole walkable area.
room1 = Polygon([(0, 0), (10, 0), (10, 10), (0, 10)])
room2 = Polygon([(15, 0), (25, 0), (25, 10), (15, 10)])
corridor = Polygon([(10, 4.5), (28, 4.5), (28, 5.5), (10, 5.5)])
area = GeometryCollection(corridor.union(room1.union(room2)))
walkable_area = pedpy.WalkableArea(area.geoms[0])
pedpy.plot_walkable_area(walkable_area=walkable_area).set_aspect("equal")
Definition of Start Positions and Exit#
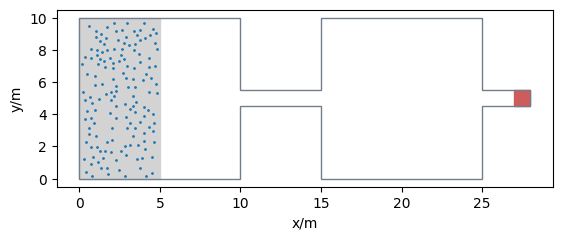
Now we define the spawning area according to RiMEA and calculate 150 positions within that area. The exit area is defined at the end of the corridor.
spawning_area = Polygon([(0, 0), (5, 0), (5, 10), (0, 10)])
num_agents = 150
pos_in_spawning_area = jps.distributions.distribute_by_number(
polygon=spawning_area,
number_of_agents=num_agents,
distance_to_agents=0.3,
distance_to_polygon=0.15,
seed=1,
)
exit_area = Polygon([(27, 4.5), (28, 4.5), (28, 5.5), (27, 5.5)])
Let’s have a look at our setup:
Show code cell source
def plot_simulation_configuration(
walkable_area, spawning_area, starting_positions, exit_area
):
axes = pedpy.plot_walkable_area(walkable_area=walkable_area)
axes.fill(*spawning_area.exterior.xy, color="lightgrey")
axes.fill(*exit_area.exterior.xy, color="indianred")
axes.scatter(*zip(*starting_positions), s=1)
axes.set_xlabel("x/m")
axes.set_ylabel("y/m")
axes.set_aspect("equal")
plot_simulation_configuration(
walkable_area, spawning_area, pos_in_spawning_area, exit_area
)
Specification of Parameters und Running the Simulation#
Now we just need to define the details of the operational model, routing and the specific agent parameters. In this example, the agents share the same parameters, ecxept for their free movement speed (and starting position).
trajectory_file = "double-botteleneck.sqlite" # output file
simulation = jps.Simulation(
model=jps.CollisionFreeSpeedModel(),
geometry=area,
trajectory_writer=jps.SqliteTrajectoryWriter(
output_file=pathlib.Path(trajectory_file)
),
)
exit_id = simulation.add_exit_stage(exit_area.exterior.coords[:-1])
journey = jps.JourneyDescription([exit_id])
journey_id = simulation.add_journey(journey)
v_distribution = normal(1.34, 0.05, num_agents)
for pos, v0 in zip(pos_in_spawning_area, v_distribution):
simulation.add_agent(
jps.CollisionFreeSpeedModelAgentParameters(
journey_id=journey_id,
stage_id=exit_id,
position=pos,
desired_speed=v0,
radius=0.15,
)
)
while simulation.agent_count() > 0:
simulation.iterate()
Visualization#
Let’s have a look at the visualization of the simulated trajectories:
from jupedsim.internal.notebook_utils import animate, read_sqlite_file
trajectory_data, walkable_area = read_sqlite_file(trajectory_file)
animate(trajectory_data, walkable_area)